It is also possible that calpain-mediated cleavage of proteins is elevated by bortezomib; this has been proposed to explain the increased degradation of IkBa caused by bortezomib high content screening treatment of various human cell lines. However, a previous peptidomics study did not detect major changes in levels of intracellular peptides when SH-SY5Y cells were treated with a calcium ionophore known to activate calpains. Another possibility is activation of autophagy by bortezomib, which is known to induce autophagy in several systems. This idea is attractive because of the large number of mitochondrial protein fragments found to be elevated by bortezomib. However, the standard marker for autophagy, LC3, showed no evidence of autophagy upon treatment of HEK293T or SH-SY5Y cells with high concentrations of bortezomib for 1 hour. Furthermore, treatment of cells with rapamycin for 1 hour to produce autophagy had little effect on the cellular peptides. Taken together, these results suggest that autophagy does not contribute to the altered peptidome observed upon treatment of cells with bortezomib for short time periods. A third possibility to explain 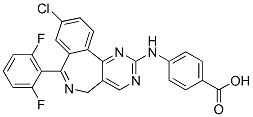 the increase in many peptides is that protein levels are induced by bortezomib. A previous RNA microarray analysis found that thousands of mRNAs were either up- or down-regulated by treatment with bortezomib for 14, 24, or 48 hours. A cross-comparison of the RNA microarray study and our results found no correlation between those proteins corresponding to up-regulated peptides and mRNA changes at 14 and 24 hour time points. Furthermore, our finding that most of the peptides were up-regulated as early as 30 minutes after the start of the exposure to bortezomib argues against a general effect on protein synthesis; while the synthesis of some proteins may be stimulated within 30 minutes of the start of bortezomib treatment, it is unlikely that all of the affected proteins will show such a rapid increase. Furthermore, a change in protein levels would not explain why all peptides derived from a particular protein are not similarly affected. For example, although many fragments of heat shock 10 kDa protein 1are elevated, one fragment is significantly decreased by 50 and 500 nM bortezomib. Similar variability in the changes of peptides derived from other proteins is observed. Therefore, an increase in the synthesis of these proteins would not explain why some of the peptides derived from these proteins were decreased by the bortezomib treatment. A fourth possibility is that bortezomib BIBW2992 interferes with the further degradation of the peptides produced by the proteasome. Although bortezomib is usually described in the literature as being highly specific for the proteasome with no off-target effects, a recent study has shown that bortezomib inhibits serine proteases such as cathepsins A and G, chymase, dipeptidyl peptidase II, and HtrA2/Omi. Although none of these enzymes are thought to function in the degradation of peptides produced by the proteasome, it is possible that bortezomib has additional off target effects and inhibits a cytosolic peptidase. This possibility would be consistent with the increase in peptides derived from cytosolic proteins as well as mitochondrial proteins; this organelle contains transporters that export peptides generated by proteases within the mitochondria, and these peptides are subsequently degraded by cytosolic peptidases. The mechanism of action of bortezomib as an antitumor agent is thought to involve a reduction in protein turnover and/or protein activation, especially for proteins such as NFkB and cyclins. To generate the active NFkB transcriptional dimeric complexes, two precursors, NFkB1and NFkB2, have to undergo limited proteolytic processing by the proteasome to yield.
the increase in many peptides is that protein levels are induced by bortezomib. A previous RNA microarray analysis found that thousands of mRNAs were either up- or down-regulated by treatment with bortezomib for 14, 24, or 48 hours. A cross-comparison of the RNA microarray study and our results found no correlation between those proteins corresponding to up-regulated peptides and mRNA changes at 14 and 24 hour time points. Furthermore, our finding that most of the peptides were up-regulated as early as 30 minutes after the start of the exposure to bortezomib argues against a general effect on protein synthesis; while the synthesis of some proteins may be stimulated within 30 minutes of the start of bortezomib treatment, it is unlikely that all of the affected proteins will show such a rapid increase. Furthermore, a change in protein levels would not explain why all peptides derived from a particular protein are not similarly affected. For example, although many fragments of heat shock 10 kDa protein 1are elevated, one fragment is significantly decreased by 50 and 500 nM bortezomib. Similar variability in the changes of peptides derived from other proteins is observed. Therefore, an increase in the synthesis of these proteins would not explain why some of the peptides derived from these proteins were decreased by the bortezomib treatment. A fourth possibility is that bortezomib BIBW2992 interferes with the further degradation of the peptides produced by the proteasome. Although bortezomib is usually described in the literature as being highly specific for the proteasome with no off-target effects, a recent study has shown that bortezomib inhibits serine proteases such as cathepsins A and G, chymase, dipeptidyl peptidase II, and HtrA2/Omi. Although none of these enzymes are thought to function in the degradation of peptides produced by the proteasome, it is possible that bortezomib has additional off target effects and inhibits a cytosolic peptidase. This possibility would be consistent with the increase in peptides derived from cytosolic proteins as well as mitochondrial proteins; this organelle contains transporters that export peptides generated by proteases within the mitochondria, and these peptides are subsequently degraded by cytosolic peptidases. The mechanism of action of bortezomib as an antitumor agent is thought to involve a reduction in protein turnover and/or protein activation, especially for proteins such as NFkB and cyclins. To generate the active NFkB transcriptional dimeric complexes, two precursors, NFkB1and NFkB2, have to undergo limited proteolytic processing by the proteasome to yield.
Month: July 2019
DN59 does not cause general disruption of cellular plasma membranes epithelial and mosquito cells did not show any toxicity at DN59 concentrations
Nor did DN59 induce substantial hemolysis of red blood cellsillustrating at concentrations as high as the 100 mM used for cryoEM. Additionally, DN59 does not inhibit the infectivity of other lipidenveloped viruses, including Sindbis virus or the negative-stranded RNA vesicular stomatitis virus. The lack of apparent disruption of cellular plasma membranes and other viral membranes may be due to lipid composition, protein incorporation, or active repair of cellular membranes. Dengue virus particles bud from 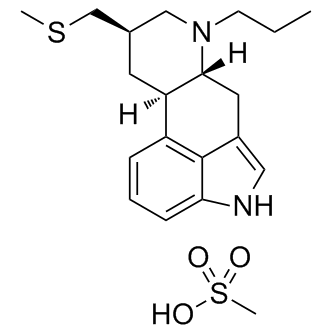 internal endoplasmic reticulum membranes of infected cells and so likely have a AB1010 different composition from the plasma membrane, although the membrane disruption activity of stem region peptides is not strongly influenced by lipid membrane composition. Schmidt et al.studied a series of similar dengue E protein stem region peptides whose sequences extensively overlap the sequence of DN59. Consistent with our earlier work, they showed that their most active peptideinhibits dengue virus infection during an entry step and can bind to synthetic lipid vesicles. Furthermore, they reported that their peptide bound to the post-fusion trimeric form of recombinant dengue surface E proteinat low pH, but did not bind to the monomeric E protein at neutral pH. They therefore proposed that the peptide neutralizes the virus by first attaching to the viral membrane, and subsequently interacting with the E post-fusion Nutlin-3 trimers that form when the virus encounters the low pH environment of the endosome, thereby preventing fusion of the virus to the endosomal membrane. Here, however, we have shown that DN59 can induce the formation of holes in the viral membrane, release the genome, and causes the viral particles to become non-infectious even before interacting with cells. The discrepancy in the mechanism of neutralization detected by our group and Schmidt et al. could possibly be due to the differences in peptide concentration used in these assays. The most likely mechanism by which DN59 or other stem region peptides can penetrate the outer layer of E glycoproteins and gain access to the virus membrane is by way of dynamic “breathing” of the virus particle. The ease with which the virus can breathe will depend on the stability of the virus, which may account in part for the differing inhibitory activities against different flaviviruses. Once the DN59 peptide has inserted itself between the E ectodomain and the membrane, it likely competes with and displaces the virus E protein stem regionfor binding to the lipid membrane and the “underside” of the E protein. Formation of holes in the viral membrane large enough for the escape of the RNA genome may involve structural changes in the surface E and M proteins, or may be due to the action of the peptide alone, similar to what is observed for some anti-microbial peptidesand what we observed with liposome vesicles. The negative charge on the tightly packaged RNA may also help the RNA to exit the virus particle once the membrane has been destabilized. Our observations show that DN59, a 33 amino acid peptide mimicking a portion of the dengue virus E protein stem region, functions through an unexpected mechanism that involves disruption of the viral membrane and release of the viral genome. Caspases are a family of cysteinyl proteases that are key mediators of apoptosis and inflammation. The apoptotic “executioner” caspasesare translated as proenzymes containing a short pro-domain, a p20 subunit, a linker region, and p10 subunit. Their canonical activation mechanism involves proteolysis by “initiator” caspasesat three distinct sites to remove the prodomain and linker region.
internal endoplasmic reticulum membranes of infected cells and so likely have a AB1010 different composition from the plasma membrane, although the membrane disruption activity of stem region peptides is not strongly influenced by lipid membrane composition. Schmidt et al.studied a series of similar dengue E protein stem region peptides whose sequences extensively overlap the sequence of DN59. Consistent with our earlier work, they showed that their most active peptideinhibits dengue virus infection during an entry step and can bind to synthetic lipid vesicles. Furthermore, they reported that their peptide bound to the post-fusion trimeric form of recombinant dengue surface E proteinat low pH, but did not bind to the monomeric E protein at neutral pH. They therefore proposed that the peptide neutralizes the virus by first attaching to the viral membrane, and subsequently interacting with the E post-fusion Nutlin-3 trimers that form when the virus encounters the low pH environment of the endosome, thereby preventing fusion of the virus to the endosomal membrane. Here, however, we have shown that DN59 can induce the formation of holes in the viral membrane, release the genome, and causes the viral particles to become non-infectious even before interacting with cells. The discrepancy in the mechanism of neutralization detected by our group and Schmidt et al. could possibly be due to the differences in peptide concentration used in these assays. The most likely mechanism by which DN59 or other stem region peptides can penetrate the outer layer of E glycoproteins and gain access to the virus membrane is by way of dynamic “breathing” of the virus particle. The ease with which the virus can breathe will depend on the stability of the virus, which may account in part for the differing inhibitory activities against different flaviviruses. Once the DN59 peptide has inserted itself between the E ectodomain and the membrane, it likely competes with and displaces the virus E protein stem regionfor binding to the lipid membrane and the “underside” of the E protein. Formation of holes in the viral membrane large enough for the escape of the RNA genome may involve structural changes in the surface E and M proteins, or may be due to the action of the peptide alone, similar to what is observed for some anti-microbial peptidesand what we observed with liposome vesicles. The negative charge on the tightly packaged RNA may also help the RNA to exit the virus particle once the membrane has been destabilized. Our observations show that DN59, a 33 amino acid peptide mimicking a portion of the dengue virus E protein stem region, functions through an unexpected mechanism that involves disruption of the viral membrane and release of the viral genome. Caspases are a family of cysteinyl proteases that are key mediators of apoptosis and inflammation. The apoptotic “executioner” caspasesare translated as proenzymes containing a short pro-domain, a p20 subunit, a linker region, and p10 subunit. Their canonical activation mechanism involves proteolysis by “initiator” caspasesat three distinct sites to remove the prodomain and linker region.
This AChE1 constraint constitutes a weakness in its adaptive capacity that might be used to develop innovative resistance
An insecticide targeting specifically the G119S AChE1 should efficiently reduce the LY2157299 frequency of the resistance allele, while the probability of OP-resistant mosquitoes developing a secondary resistance to the new insecticide is predicted to be quite low. To address the feasibility of this approach, compounds were screened for their capacity to inhibit more efficiently the G119S-substitutedAChE1 than the wild typeAChE1. Further biochemical analysis, bioassays on mosquito larvae from susceptible and resistant A. gambiae and C. pipiens strains sharing the same genetic background, as well as gene population modeling show that application of compounds with such properties is predicted to rapidly restore OP susceptibility in field populations. We show here that PTFs qualify as a new class of reversible and competitive AChE1 inhibitors, with preferential efficacy toward OP-insensitive AChE1 in vitro and OP-resistant C. pipiens or A. gambiae mosquito larvae in vivo. The fact that PTFs behave as reversible and competitive AChE1 inhibitors strongly suggests that they target the docking sites for acetylcholine, i.e. the peripheralor the activesites. Furthermore, PTFs preferentially inhibit all OP-insensitive mutants, whose substituted positions line the A-site and are thought to confer OP-insensitivity by steric hindrance. It is therefore unlikely that PTFs can freely enter the A-site of OP-insensitive enzymes and a more likely hypothesis is that PTFs bind on top of the A- or the P-site, thereby blocking access to the substrate to either site. This agrees with the absence of correlation between PTF efficacy and specific chemical groups at the R1,R 2 or R3 positions, suggesting that docking of the core PTF structure to its target might be stabilized through multiple interactions with neighboring residues. Availability of AChE1 three-dimensional structure, alone or in complex with PTFs, should help understand the structural basis of the interaction. PTFs proved to be efficient in vivo on C. pipiens and A. gambiae larvae and showed similar selectivity toward OP-resistant larvae, although with different RLD50 values. Behavioral or physiological differences might account for this difference; Anopheles gambiae larvae are surface filter-feeders while C. pipiens are deeper filterfeeders that regularly swim up to breathe. Previous work also demonstrated that the dominance of resistance in C. pipiens varies with environment, such as food, water quality and  shape of the cups used in bioassays. All these parameters might impact on insecticide uptake or induced mortality. Since PTFs inhibit all types of OP-insensitive AChE1 in vitro, i.e. G119S-, F290V- or F331W-substituted, this strongly suggests that PTF molecules might exhibit a broad larvicidal spectrum toward most OP-resistant field populations. In conclusion, this study validates an innovative approach for resistance management of mosquito vectors, based on the development of molecules targeting preferentially enzymes already insensitive to MK-1775 955365-80-7 currently used insecticides. PTFs identified here behave as preferential inhibitors of AChE1 mutants insensitive to OP and CX and as preferential killers of OPresistant C. pipiens and A. gambiae larvae in bioassays. This approach should allow both efficient vector and resistance control management: The preferential killing of resistant mosquitoes mimics a situation of negative cross-resistance, in which frequency of resistance alleles decreases much faster than when insecticides acting on other targets are used. Furthermore, since wild type OP-susceptible alleles are, by construction, PTFresistant alleles, odds are against the selection of other resistance events.
shape of the cups used in bioassays. All these parameters might impact on insecticide uptake or induced mortality. Since PTFs inhibit all types of OP-insensitive AChE1 in vitro, i.e. G119S-, F290V- or F331W-substituted, this strongly suggests that PTF molecules might exhibit a broad larvicidal spectrum toward most OP-resistant field populations. In conclusion, this study validates an innovative approach for resistance management of mosquito vectors, based on the development of molecules targeting preferentially enzymes already insensitive to MK-1775 955365-80-7 currently used insecticides. PTFs identified here behave as preferential inhibitors of AChE1 mutants insensitive to OP and CX and as preferential killers of OPresistant C. pipiens and A. gambiae larvae in bioassays. This approach should allow both efficient vector and resistance control management: The preferential killing of resistant mosquitoes mimics a situation of negative cross-resistance, in which frequency of resistance alleles decreases much faster than when insecticides acting on other targets are used. Furthermore, since wild type OP-susceptible alleles are, by construction, PTFresistant alleles, odds are against the selection of other resistance events.
The HA-mediated entry assay indicated that the tropism of H7N9 was quite similar to avian H5N1
Until now, information is still limited about the transmission and Compound Library pathogenicity associated with this novel avian influenza H7N9 virus. Currently, the NA inhibitors Oseltamivir and Zanamivir are the front-line therapeutic options against this novel H7N9 influenza virus, which contains an S31N mutation in its M2 protein and thus confers resistance to the M2 ion channel blockers. However, recent studies have reported that H7N9 isolates with the R292K mutation are resistant to Zanamivir, Peramivir and Oseltamivir. Two of 14 patients infected with the novel H7N9 influenza virus possessing an R292K NA mutation had a poor clinical outcome. Therefore, discovering novel antiviral targets and drug candidates is urgently expected. HA-mediated virus-host membrane fusion is a prerequisite for the viral life cycle and is a promising antiviral target. Based on the HA-dependent membrane fusion model, we designed a cholesterol-conjugated peptide, named P155-185-chol, which is derived from the amino acid 155-185 region of H7N9 HA2. P155-185-chol demonstrated H7N9pp-specific inhibition of infection and exhibited significant inhibitory effects against H7N9 virus. In summary, our study demonstrated that the established H7N9-pseudotyped particle system can largely mimic the entry properties of the H7N9 virus, thus providing a relatively safe experimental system for studying the entry of H7N9 virus and for high-throughput screening of entry inhibitors, such as small molecules, peptides or antibodies. A potent anti-H7N9 peptide was successfully identified in this study. When used in combination with neuraminidase  inhibitors, P155-185-chol showed additive and significant inhibitory effects against H7N9. By virtue of its distinct mechanisms of inhibition, P155-185-chol may be further optimized to improve its activity, highlighting a potential therapeutic candidate for H7N9 infections that can be used in combination with other anti-flu drugs. Obesity is a high risk factor for the development of many human pathologies, including insulin resistance, type 2 diabetes, hyperlipidemia, hypertension, cardiovascular disease, and cancer. Mounting evidence RWJ 64809 suggests that obesity is a multi-factorial disorder caused by a variety of genetic and endocrine abnormalities, some medicines, a low metabolic rate, nutritional and environmental factors, as well as imbalance of energy homeostasis. A number of studies have demonstrated that adipose tissue plays a critical role in the regulation of energy metabolism by secreting adipokines. However, there is strong evidence suggesting that abnormal expansion/accumulation of adipose tissue, which is largely associated with excessive adipocyte differentiation, increased numbers and lipid contents of fat cells, are closely linked to the development of obesity. Thus, any compound that inhibits excessive adipocyte differentiation and/or adipocyte hyperplasia/hypertrophy may have preventive and/or therapeutic potential against obesity and obesity-related disease. Research has accumulated to indicate that the differentiation of preadipocyte into adipocyte is controlled by a complex network of a variety of cellular proteins, including transcription factors, cell cycle-related proteins, adipocyte-specific genes, lipogenic enzymes, and signaling proteins. 3T3-L1 is a well-established murine preadipocyte cell line and has widely been used for understanding the molecular regulation of adipocyte differentiation and for screening potential anti-obesity drugs or agents. Interestingly, recent studies have shown that a number of polyphenolic natural products inhibit adipogenesis and have anti-proliferative and/or proapoptotic effects on 3T3-L1 adipocytes at high concentrations.
inhibitors, P155-185-chol showed additive and significant inhibitory effects against H7N9. By virtue of its distinct mechanisms of inhibition, P155-185-chol may be further optimized to improve its activity, highlighting a potential therapeutic candidate for H7N9 infections that can be used in combination with other anti-flu drugs. Obesity is a high risk factor for the development of many human pathologies, including insulin resistance, type 2 diabetes, hyperlipidemia, hypertension, cardiovascular disease, and cancer. Mounting evidence RWJ 64809 suggests that obesity is a multi-factorial disorder caused by a variety of genetic and endocrine abnormalities, some medicines, a low metabolic rate, nutritional and environmental factors, as well as imbalance of energy homeostasis. A number of studies have demonstrated that adipose tissue plays a critical role in the regulation of energy metabolism by secreting adipokines. However, there is strong evidence suggesting that abnormal expansion/accumulation of adipose tissue, which is largely associated with excessive adipocyte differentiation, increased numbers and lipid contents of fat cells, are closely linked to the development of obesity. Thus, any compound that inhibits excessive adipocyte differentiation and/or adipocyte hyperplasia/hypertrophy may have preventive and/or therapeutic potential against obesity and obesity-related disease. Research has accumulated to indicate that the differentiation of preadipocyte into adipocyte is controlled by a complex network of a variety of cellular proteins, including transcription factors, cell cycle-related proteins, adipocyte-specific genes, lipogenic enzymes, and signaling proteins. 3T3-L1 is a well-established murine preadipocyte cell line and has widely been used for understanding the molecular regulation of adipocyte differentiation and for screening potential anti-obesity drugs or agents. Interestingly, recent studies have shown that a number of polyphenolic natural products inhibit adipogenesis and have anti-proliferative and/or proapoptotic effects on 3T3-L1 adipocytes at high concentrations.
Our observation that bortezomib is a weak inhibitor of aminopeptidase activity in HEK293T cells was initially considered
Increasing evidence suggests that some proteasome inhibitors exibit an allosteric effect on proteasome stability; MG262 treated purified 26S proteasomes were resistant to apyrase-induced proteasome dissociation whereas MG132 had no effect on proteasome stability. In other studies, bortezomib was reported to activate the beta 2 subunit, which cleaves at basic amino acids. The previous peptidomic study with epoxomicin noted that many of the peptides which were elevated by this compound contained an acidic residue in the P1 position of the cleavage site required to produce these peptides. Because epoxomicin does not inhibit the beta 1 AG-013736 319460-85-0 subunit responsible for cleavage at acidic residues, it would be expected that inhibition of the beta 2 and beta 5 subunits would lead to a greater share of protein degradation occurring at acidic residues. However, some of the peptides that were elevated upon treatment of cells with epoxomicin, and most of the peptides elevated upon treatment of cells with bortezomib, have hydrophobic residues in the P1 position of the cleavage site. Similarly, carfilzomib and MG262 also elevated levels of peptides that required cleavage at hydrophobic sites; all of these inhibitors are most potent at the beta 5 subunit, which is responsible for cleaving at hydrophobic sites. Somehow the inhibitors of the beta 5 subunit appear to be activating the beta 5 subunit, possibly by affecting the opening of the gate within the 20S proteasome core particle; bortezomib, MG262, and epoxomicin were all found to open this  gate. In the present study, we found that bortezomib showed EX 527 comparable inhibition of the 20S core particle and an open-gate mutant of this 20S core particle when assayed with the standard substrate for beta 5 activity, but it is possible that allosteric regulation of the proteasome affects the intracellular peptides differently than the synthetic substrate. For example, Kisselev et al found that hydrophobic peptides including SuccLeu-Leu-Val-Tyr-AMC can trigger gate opening and stimulate the activity of 20S particles. A related possibility is that the various proteasome forms are differentially affected by inhibitors. In support of this hypothesis, the antiviral drug ritonavir was found to activate the chymotryptic-like activity of the 26S form of the proteasome while inhibiting the 20S form. Although we found no difference in the effect of bortezomib on the chymotryptic-like activity of the 26S versus the 20S form, or the 20S form activated by Blm10, it remains possible that allosteric effects of the proteasome inhibitors influence cleavage of the intracellular peptides by the various proteasome forms. Our results do not support the hypothesis that the proteasome inhibitors have off-target effects on enzymes that further degrade the peptides produced by the proteasome. A number of different enzymes have been implicated in the degradation of proteasome products, including oligopeptidases, aminopeptidases, and TPP2. Based on a bioinformatic approach, it was proposed that bortezomib could be an inhibitor of TPP2, although no direct evidence of this was provided. In the present study, we could not detect any inhibition of TPP2 activity by bortezomib. Furthermore, peptidomic analysis of cells treated with butabindide, a potent and selective TPP2 inhibitor, did not produce dramatic changes in the cellular peptidome. These results suggest that the bortezomib-induced changes in the cellular peptidome are not due to inhibition of TPP2. The failure of butabindide to cause large changes in the cellular peptidome suggests that TPP2 does not play a major role in the degradation of the intracellular peptides detected with the peptidomic technique, consistent with the finding that TPP2 is not required for the production of peptides bound to HLA.
gate. In the present study, we found that bortezomib showed EX 527 comparable inhibition of the 20S core particle and an open-gate mutant of this 20S core particle when assayed with the standard substrate for beta 5 activity, but it is possible that allosteric regulation of the proteasome affects the intracellular peptides differently than the synthetic substrate. For example, Kisselev et al found that hydrophobic peptides including SuccLeu-Leu-Val-Tyr-AMC can trigger gate opening and stimulate the activity of 20S particles. A related possibility is that the various proteasome forms are differentially affected by inhibitors. In support of this hypothesis, the antiviral drug ritonavir was found to activate the chymotryptic-like activity of the 26S form of the proteasome while inhibiting the 20S form. Although we found no difference in the effect of bortezomib on the chymotryptic-like activity of the 26S versus the 20S form, or the 20S form activated by Blm10, it remains possible that allosteric effects of the proteasome inhibitors influence cleavage of the intracellular peptides by the various proteasome forms. Our results do not support the hypothesis that the proteasome inhibitors have off-target effects on enzymes that further degrade the peptides produced by the proteasome. A number of different enzymes have been implicated in the degradation of proteasome products, including oligopeptidases, aminopeptidases, and TPP2. Based on a bioinformatic approach, it was proposed that bortezomib could be an inhibitor of TPP2, although no direct evidence of this was provided. In the present study, we could not detect any inhibition of TPP2 activity by bortezomib. Furthermore, peptidomic analysis of cells treated with butabindide, a potent and selective TPP2 inhibitor, did not produce dramatic changes in the cellular peptidome. These results suggest that the bortezomib-induced changes in the cellular peptidome are not due to inhibition of TPP2. The failure of butabindide to cause large changes in the cellular peptidome suggests that TPP2 does not play a major role in the degradation of the intracellular peptides detected with the peptidomic technique, consistent with the finding that TPP2 is not required for the production of peptides bound to HLA.